Understanding plant genomes is crucial for plant breeding, agricultural productivity, and sustainability. However, plant genomes are diverse in size, composition, and complexity, experimentally generating analogous genomic resources for hundreds of thousands of plant species is challenging and impractical. This highlights the need for developing cross-species models capable of capturing evolutionary conservation across diverse plant species.
Supervised sequence models are successful in understanding DNA sequence function but typically require large-scale labeled data, such as ENCODE-scale datasets, to achieve robust performance. Such extensive labeled data is often scarce in plant genomics. However, the success of self-supervised language models (LMs) offers a promising alternative. In this paradigm, a base model is pre-trained on vast amounts of unlabeled biological sequences to learn evolutionary conservation. Pre-trained models are then fine-tuned on limited labeled data, enabling better performance on downstream tasks and enhancing generalizability across species relative to existing methods.
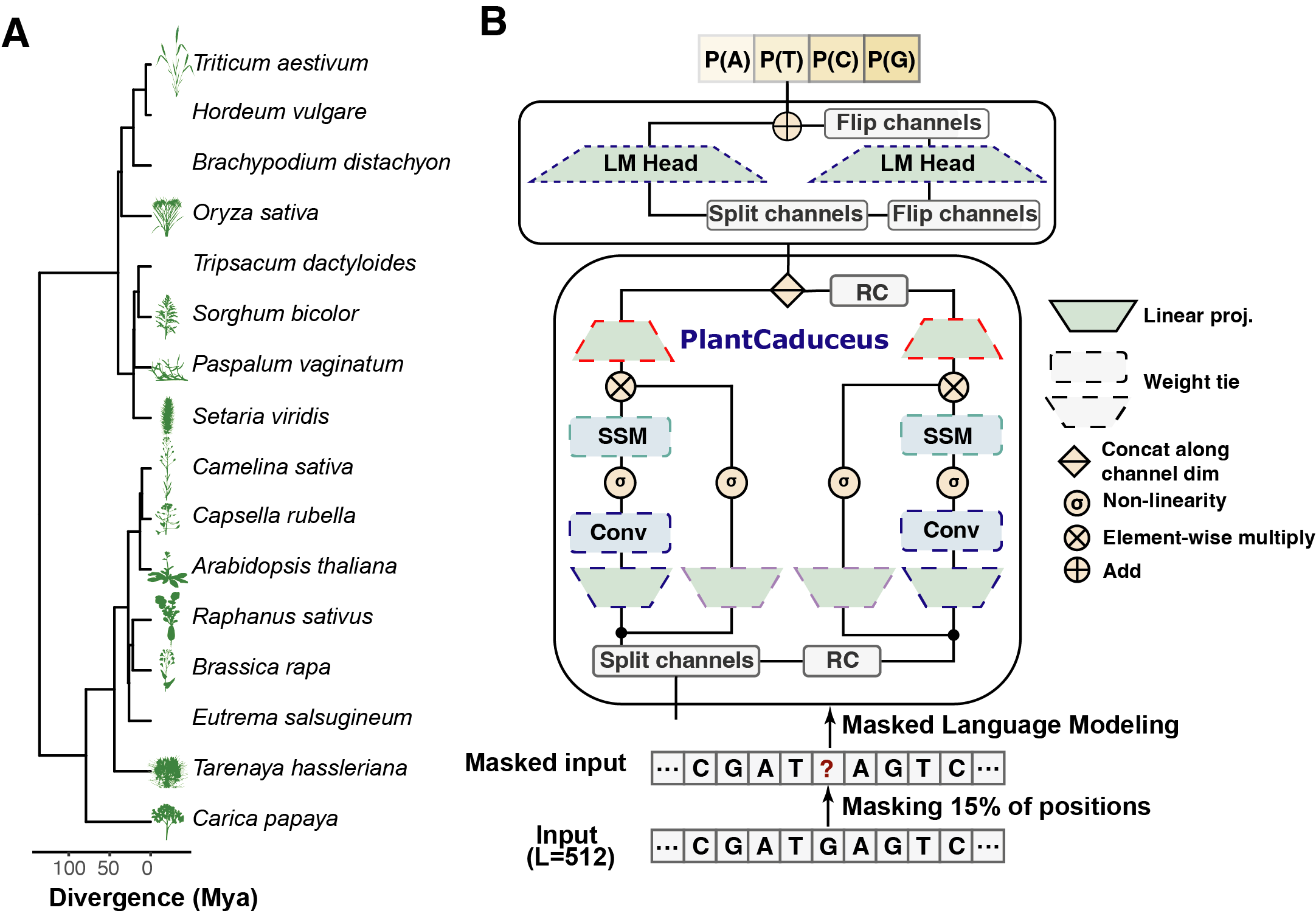
PlantCaduceus (PlantCAD) builds upon Caduceus (Schiff et al., 2024) and Mamba (Gu et al., 2023) architectures to model diverse genomes at single nucleotide resolution. PlantCaduceus is trained on 16 Angiosperm genomes spanning 160 million years of evolutionary history, to capture evolutionary conservation and variation across species. PlantCaduceus takes 512 base pair (bp) windows of input sequences, tokenizing them into single nucleotides, and is pre-trained using a masked language modeling objective.
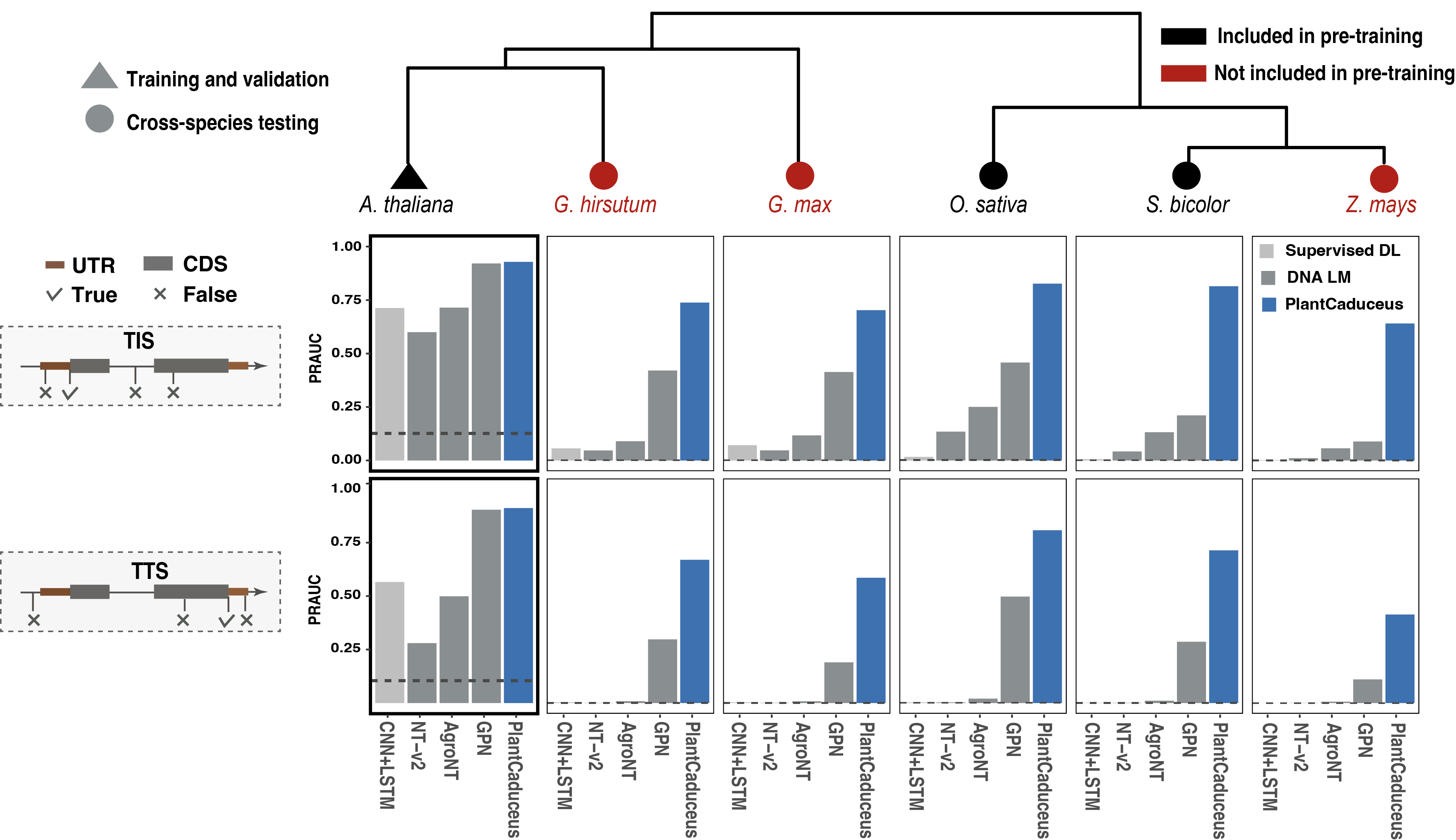
Accurate prediction of translation initiation site (TIS), translation termination site (TTS), and splice donor and acceptor sites, is crucial in gene annotation. To explore PlantCaduceus's performance on gene annotation tasks, we generated training and validation datasets for these four tasks, using Arabidopsis thaliana TAIR10, a relatively well-annotated genome released over two decades ago. To evaluate the cross-species prediction performance, we generated three extremely imbalanced cross-species testing datasets for rice, sorghum, and maize. With only training on Arabidopsis, PlantCaduceus outperformed all benchmark models on TIS and TTS tasks, outperforming the best benchmark model GPN by 7.23-fold and 3.75-fold for TIS and TTS when transfered to 160 million year diverged maize, respectively.
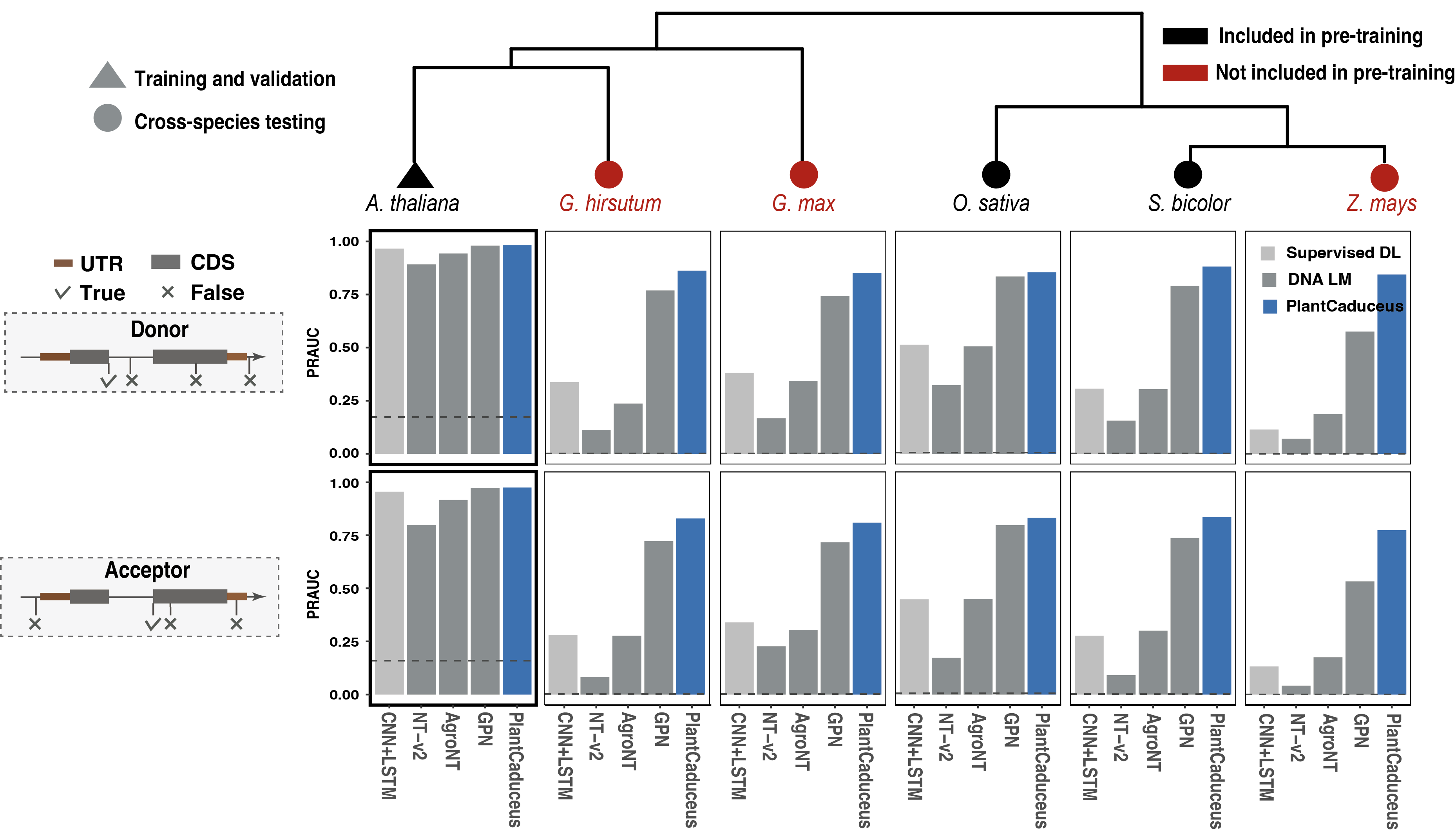
With similar way as TIS and TTS, we generated training, validation and testing datasets for splice donor and acceptor tasks. We found PlantCaduceus outperformed all benchmark models on splice donor and acceptor sites tasks, outperforming the best benchmark model GPN by 1.47-fold and 1.45-fold for splice donor and acceptor sites, respectively.
The training objective of PlantCaduceus is to predict masked nucleotides based on sequence context; if a pre-trained multi-species DNA LM can accurately predict masked tokens, it suggests that similar sequence patterns, conserved across different species, were frequently observed during pre-training. We hypothesize that the predicted likelihood of the reference allele versus the alternate allele can identify deleterious mutations, as mutations in conserved regions across species are likely deleterious.
Deleterious mutations tend to have lower frequencies within a population due to selective constraints, we therefore used minor allele frequency (MAF) to quantify the deleteriousness of mutations predicted by different methods. Despite the potential for low MAF in neutral/beneficial alleles, we believe this approach provides useful signals for assessing deleterious mutations. Both phyloP and phastCons assess evolutionary constraint using multiple sequence alignments and phylogenetic models, assigning higher scores to conserved regions. We found the deleterious mutations identified with the zero-shot strategy of PlantCaduceus show a three-fold enrichment of rare alleles compared to phastCons. For missense mutations, PlantCaduceus matches the performance of the state-of-the-art protein LM.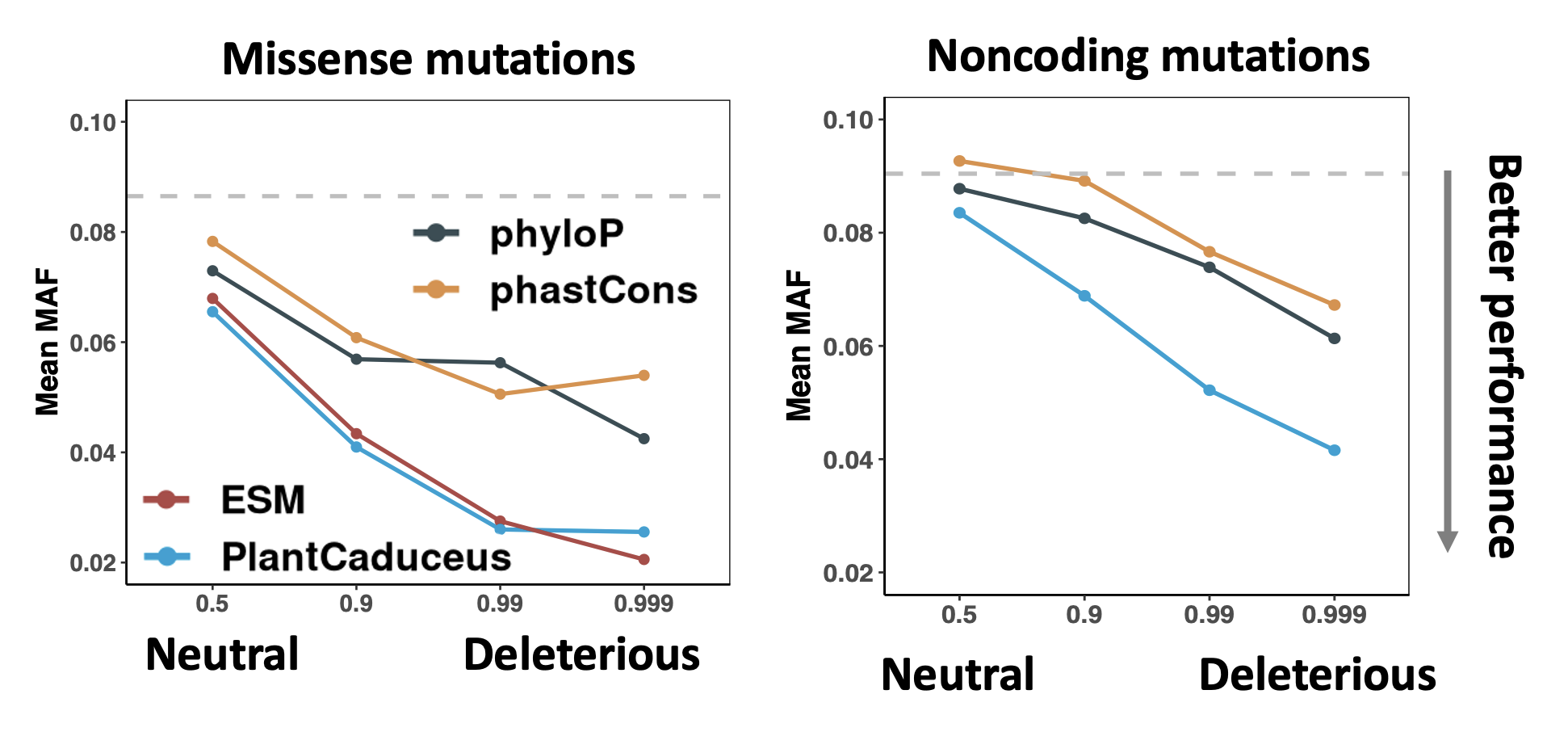
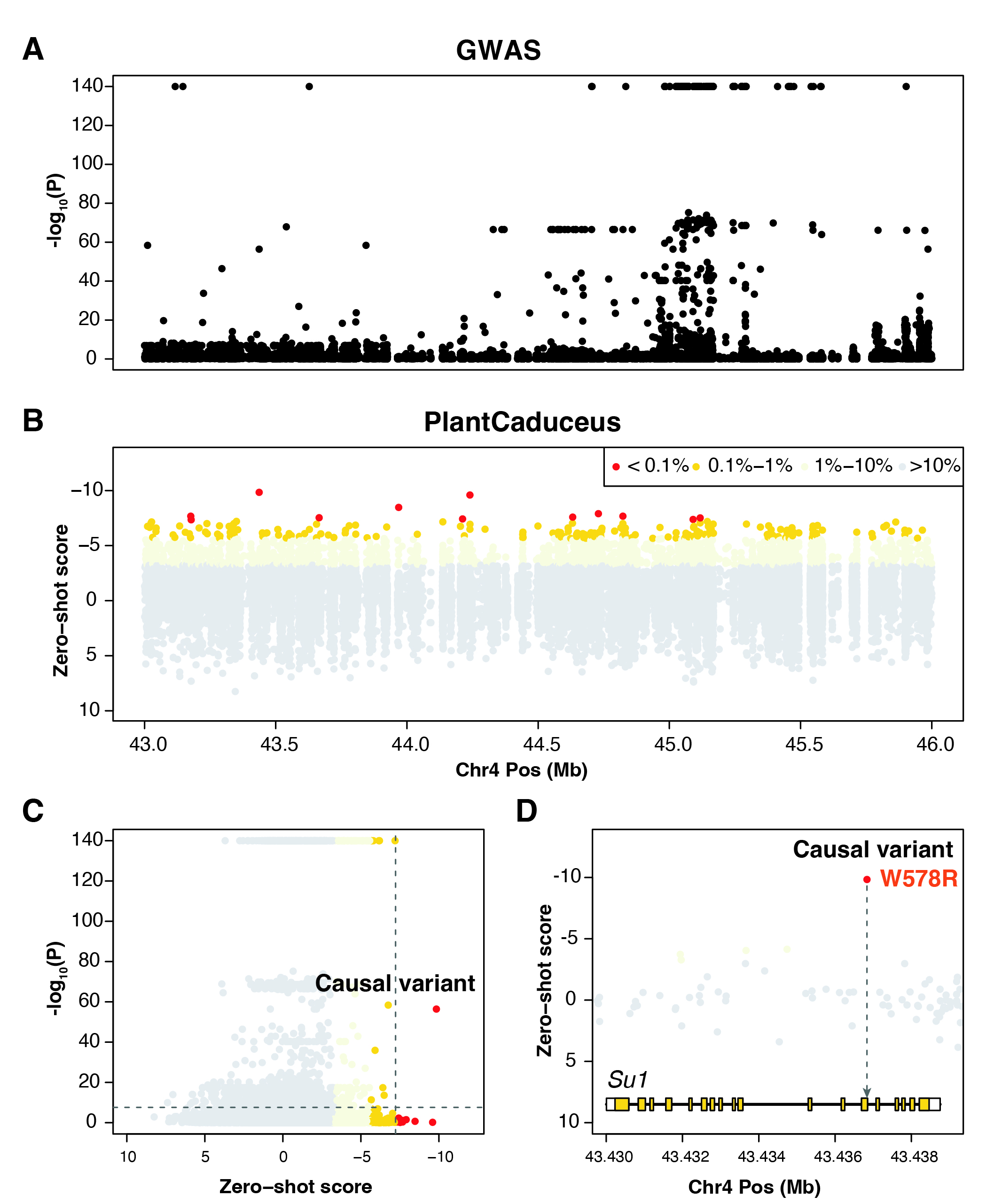
Sweet corn, a popular vegetable variant of maize, owes its characteristic sweetness to a mutation at the sugary1 (Su1) locus, specifically the W578R mutation (Tracy et al., 2006). This mutation disrupts starch metabolism, leading to the accumulation of phytoglycogen, which gives sweet corn its creamy texture. Although GWAS results revealed numerous significant peaks on chromosome 4, identifying the exact causal mutations is challenging due to linkage disequilibrium (Panel A). By integrating zero-shot scores from PlantCaduceus with GWAS data, we successfully pinpointed the W578R mutation in the Su1 region as the causal variant.
We introduced PlantCaduceus, a multi-species plant DNA LM pretrained on a curated set of 16 evolutionarily distant Angiosperm genomes, enabling cross-species prediction of functional annotations with limited data. PlantCaduceus leverages Mamba and Caduceus architectures to support bi-directional, reverse complement equivariant sequence modeling. We demonstrated the superior cross-species performance of PlantCaduceus on five tasks involving transcription, translation, and evolutionary constraint modeling. These results highlight the potential of PlantCaduceus to serve as a foundational model for comprehensively understanding plant genomes.
@article{zhai2025plantcaduceus,
author = {Zhai, Jingjing and Gokaslan, Aaron and Schiff, Yair and Berthel, Ana and Liu, Zong-Yan and Lai, Wei-Yun and Miller, Zachary R and Scheben, Armin and Stitzer, Michelle C and Romay, Cinta and Buckler, Edward S. and Kuleshov, Volodymyr},
title = {PlantCaduceus: Cross-species modeling of plant genomes at single nucleotide resolution},
journal = {Proceedings of the National Academy of Sciences},
volume = {},
number = {},
pages = {},
year = {2025},
doi = {10.1073/pnas.2421738122},
URL = {https://doi.org/10.1073/pnas.2421738122}
}